Sequencing-based monitoring of pathogenic viruses in wastewater
DOWNLOAD THE PAPER
Graphical Abstract
The concept of wastewater-based epidemiology (WBE), which was initially developed for polioviruses (and a few other infections) many years ago, was widely used during the COVID-19 epidemic. In contrast to traditional methods, WBE offers low-cost, immediate, and objective findings. It is based on extracting, discovering, examining, and elucidating chemical or biological components, often known as biomarkers, from a wastewater matrix.
Nowadays, wastewater surveillance has become an important tool for monitoring the spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus within communities. The accurate, sensitive, and high-throughput detection of SARS-CoV-2 in wastewater is of great importance for WBE. Researchers found Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR) has a fast and specific amplification ability of target DNA/RNA sequence. As a result, it has been widely used in the detection and quantification of human pathogens (SARS-CoV-2) in wastewater, but it also has some drawbacks. The limitation of RT-qPCR is its poor throughput, which only allows for the simultaneous analysis of a few genomic targets. The limitation of PCR-based approaches is that the pathogen genome must be known in advance to construct the primer and probe. Additionally, it is challenging to distinguish between all variants present in the sample by focusing only on a few mutations, and in fact, they are constrained by the number of targets that may be multiplexed per reaction. On the other hand, Nanopore sequencing is a different method compared to widely used qPCR in its capacity to detect a part of or the whole viral genomes. It is also very powerful in detecting the diverse viral genomes in environmental samples. It allows for the full screening of all probable mutations without the need for prior knowledge and has thus been widely used to characterise diseases and viruses.
Oxford Nanopore Technology (ONT), as a long-read technology, offers improved phasing of polymorphic genes, detection and appropriate characterisation of structural rearrangements, real-time data collecting, and shorter turnaround times than short-read methods. Therefore, the objective of this research is to use Nanopore Targeted Sequencing (NTS) to develop quantitative detection of SARS-CoV-2, and RT-qPCR will be used as a validation of sequencing-based data. In addition, NTS will be further developed for multiple virus detection (Influenza A&B, adenovirus, and rhinovirus). With the development of COVID-19, it becomes more important to detect the SARS-CoV-2 variants in wastewater due to the ongoing mutation of the virus and its threat to public health. This research will also investigate the sequencing-based monitoring of SARS-CoV-2 variants in wastewater. Overall, this study will help to identify a highly sensitive, high-throughput tool for detecting and quantifying SARS-CoV-2 variants and other selected pathogenic viruses in wastewater.
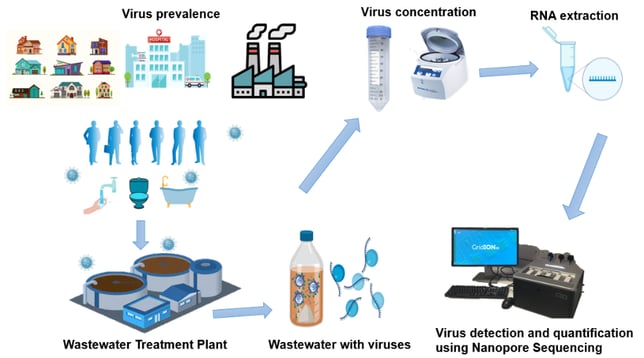
Figure 1 – Graphical representation of virus detection and quantification process using Nanopore sequencing
Author
Student name: Tanjila Alam Prosun
Affiliation: University of Wollongong, School of Civil, Mining, and Environmental Engineering.
Supervisors: Principal supervisor- Associate professor Guangming Jiang
Co-supervisor- Professor Faisal I. Hai
The research will be completed by the middle of 2025. I am working with wastewater (Influent) from the wastewater treatment plants (WWTPs). This project will help to identify the community’s health condition and the virus prevalence in the water as well. That’s why it is directly related to the water industry.
